Cross-kingdom RNA interference (RNAi) in in pathogen control
Bernhard Timo Werner
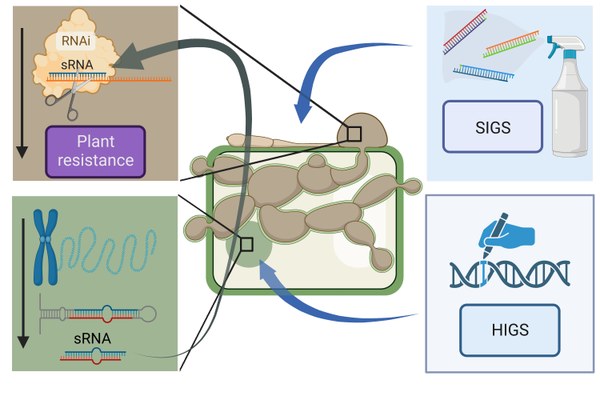
Background: Plants are known to produce non-coding small regulatory RNAs (sRNAs) with anti-fungal properties. These sRNAs target fungal genes via the mechanism of cross kingdom RNAi (ckRNAi) leading to a reduction of pathogen growth and disease severity. The validity of this concept was, for instance, proven for the fungal pathogen Verticillium dahliae, which causes devastating diseases with yield losses of up to 35% in more than 200 other plant species. Moreover, sRNAs such as homologs of cotton microRNA-159 are conserved and found in all land plants, and effectively inhibit fungal growth by silencing the isotrichodermin C-15 hydroxylase gene of V. dahliae. While, designed sRNAs are already used routinely in research and commercial settings to fight plant diseases, by either expressing them via transgenic approaches in the plant itself by host-induced gene silencing (HIGS) or by applying sRNAs onto leaves (spray-induced gene silencing, SIGS), the potential of naturally occurring sRNAs remains untapped.
Project: To exploit this untapped resource, we aspire to identify wheat sRNAs with anti-fungal properties against the notorious pathogens Fusarium graminearum, Pyricularia oryzae and Puccinia striiformis. By employing deep sequencing of the transcriptome, degradome and sRNA-ome the aim is to dissect ckRNAi events in order to find resistance-promoting sRNAs. Functionally validated sRNAs will represent candidates for engineering a new generation of inducible artificial trans-acting small-interfering RNA (tasiRNA) vectors with a broad resistance activity against several fungal pathogens. Utilizing naturally occurring sRNAs in plant protection has a huge potential for the development of bio-control agents conferring more durable resistances without undesirable off-target effects on the environment.
Lab tools / techniques: ckRNAi-seq, stem loop PCR, syn-tasiRNA, RLM-RACE, host-/spray-induced gene silencing.