Regulatory coordination of root immunity at cell type resolution
Ruth Schäfer
Collaborators: Sascha Ott (University of Warwick, UK), Cécile Raynaud (Université Paris-Saclay, France)
Background: Plant roots present an almost unexplored source for enhancing plant stress resilience. Roots show a concentric organisation with epidermis, cortex, endodermis, pericycle and stele (e.g. phloem, xylem) as principal cell types. This organisation reflects partitioning of root function by cell types which provides a flexibility in overall root functionality under changing environments. While individual cell types differently contribute to abiotic stress signalling their function in immunity (defence against pathogens) is mostly unknown. In addition, the activation of immunity can interfere with plant development. Understanding the contribution of cell types to overall immunity is essential for the generation of crops with a superior fitness under abiotic and biotic stress.
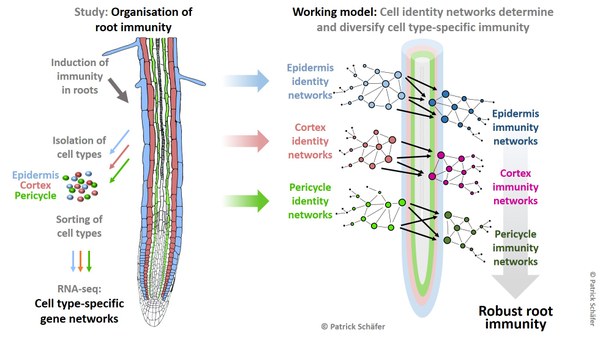
Project: To understand the organisation of immunity in root cell types, we have analysed the transcriptional networks in epidermis, cortex and pericycle cells of Arabidopsis roots treated with two known elicitors of immunity - bacterial flagellin (flg22) and the plant-derived Pep1. Our studies revealed that both elicitors activated distinct immunity networks in each cell type. To identify underlying regulatory principles, we developed a combinatorial promoter analysis tool for high accuracy prediction of functional promoter motifs. This analysis revealed pairs of transcription factors (TFs) to regulate cell type-specific immunity. Moreover, cell identity specifies cell type-specific immunity to accurately link immune response to the functional capabilities of each cell type. We want to understand how cell identity and cell age affect stress adaptation in parallel to developmental processes in individual cell types and to determine the regulatory role of specific TF pair combinations therein.
Lab tools / techniques: Cell type-specific and single cell RNAseq/transcriptomics, FACS, combinatorial promoter analytics, cell cycle/growth and immunity assays, CLSM, protein interaction assays
Relevant publications:
Üstüner, S., Schäfer, P., & Eichmann, R. (2022) Development specifies, diversifies and empowers root immunity. EMBO Reports: 23: e55631.
Rich-Griffin, C., Eichmann, R., Reitz, M. U., Hermann, S., Woolley-Allen, K., Brown, P. E., Wiwatdirekkul, K., Esteban, E., Pasha, A., Kogel, K. H., Provart, N. J., Ott, S., & Schäfer, P. (2020) Regulation of Cell Type-Specific Immunity Networks in Arabidopsis Roots. Plant Cell 32: 2742–2762.
Rich-Griffin, C., Stechemesser, A.H., Finch, J., Lucas, E.S., Ott, S., Schäfer, P. (2020) Single-cell transcriptomics - a high-resolution avenue for plant functional genomics. Trends in Plant Science 25:186-197.
Lagunas, B., Achom, M., Bonyadi-Pour, R., Pardal, A.J., Richmond, B.L., Sergaki, C., Vázquez, S., Schäfer, P., Ott, S., Hammond, J.P, Gifford, M.L (2019) Regulation of resource partitioning coordinates nitrogen and rhizobia responses and autoregulation of nodulation in the legume Medicago truncatula. Molecular Plant 12: 833-846.
Eichmann, R., Schäfer, P. (2015) Growth versus immunity-a redirection of the cell cycle? Current Opinion in Plant Biology 26:106-112.
