Research Summary
Chromatin Changes in Differentiation and Malignancies
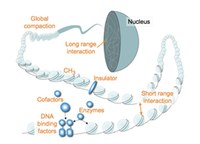
The differentiation of cells during normal development and disease is largely governed by changes to chromatin structure that manifest at several levels to determine gene activity. At a more local level, genes are embedded in active or in repressive chromatin structures. Changes between these states are modulated and shaped by DNA binding transcription factors recruiting histone modifying and nucleosome remodelling enzymes to chromatin. At a more global level, structural proteins drive chromatin folding and loop formation and establish chromosomal contacts to regulate larger genomic regions. Local and global chromatin regulation are tightly interconnected.
Since 2010 the TRR81 has made substantial progress in our understanding of how pathways transmitting developmental signals interface with histone modifying enzymes to change chromatin structure during muscle and neuronal differentiation. We unraveled molecular functions of ATP-dependent nucleosome remodellers in diverse developmental contexts, including heart development and spermatogenesis. In addition, factors and mechanisms driving chromosome conformation, topologically associated domains and interactions between enhancers and promoters, were uncovered in different cell systems. Finally, our understanding of the molecular mechanisms underlying global chromatin compaction during X-chromosome inactivation and spermatogenesis has been significantly advanced.
The focus of the current funding period (2018-2022) is the utilization of new and refined methods allowing “Chromatin Changes in Differentiation and Malignancies” to be addressed at unprecedented levels of detail. Among these are the development of small molecules to modulate chromatin regulators. Improved CRISPR methods that allow for precise changes of genomic DNA and specific targeting of chromatin regulators to specific genomic loci. Novel next generation sequencing approaches have been introduced, some of which invented by TRR81 members, to probe chromatin contacts over large distances. Sensitivity and resolution of key techniques have been dramatically improved such that analyses of the epigenomes of small numbers or even single cells have become possible. Finally, super resolution microscopy is continuously being refined by the TRR81 and will be utilised.
One of the strengths of the TRR81 is to apply these techniques to multiple, complementary models to investigate normal and pathological mechanisms that lead to chromatin changes. The differentiation models include spermatogenesis, immune cells and their activation by signalling, hematopoietic stem cells and their differentiation, ES cells and cell lines that allow differentiation towards neural and muscle lineages, cerebral organoid cultures and cancer cells. Disease models include tumor formation, neurodevelopmental syndromes, inflammation, muscle dystrophy and hemoglobinopathies.
The TRR81 will advance our understanding of epigenetic mechanisms underlying differentiation in healthy and in diseased cells and will provide the basis for the future development of novel therapeutic approaches.
Cover art: BBA-Molecular Cell Research, Vol. 1783, Issue 11