WASP
Single cell RNA-seq (scRNA-seq) enables analysis of cellular transcriptomes in an unprecedented resolution, allowing e.g., the identification of previously undiscovered rare cell populations with corresponding marker genes, detection of heterogeneity between cells of the same type or to follow transcriptional programs of cells during differentiation. Current high-throughput sequencing techniques yield enormous amounts of data that need to be analyzed. Hence, there is need for bioinformatic analysis solutions adapted to the specific challenges deriving from scRNA-seq data.
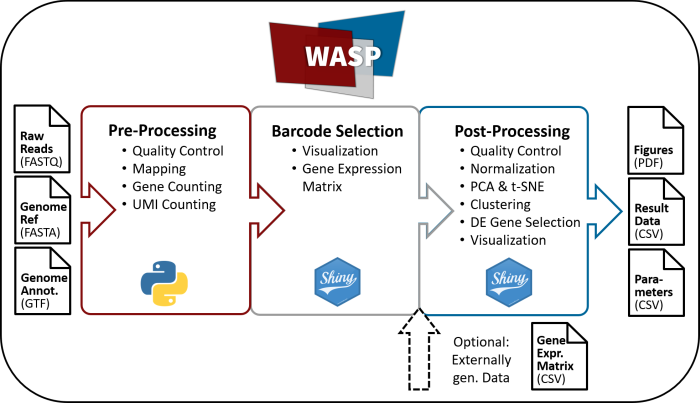
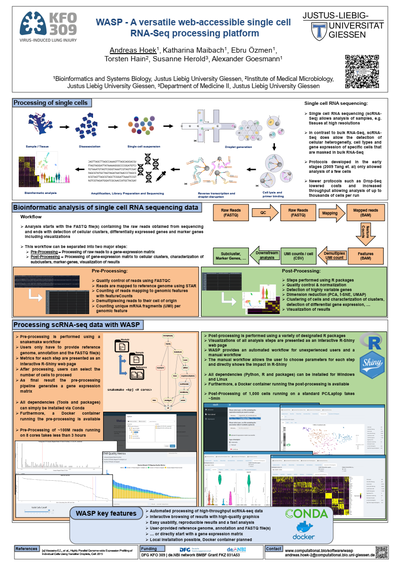
Here we present WASP - a versatile, web-accessible single cell RNA-Seq processing platform designed for the management, analysis and interpretation of scRNA-seq high-throughput data. The software addresses all aspects from initial quality control, demultiplexing and reference alignment to downstream statistical evaluation. Furthermore, it provides an automated workflow suitable for both non-bioinformaticians as well as experts. The software is available via a web-based interface and supports deployment to local as well as cloud-based compute infrastructures. For an easy installation, WASP can be used via Docker, Conda and as Windows standalone version.
Finally, the application has already successfully been applied to various data sets derived from, among others, mouse lung organoid cells.
The software is currently under peer review.
For further information & usage instructions check out our Github & user manual or contact us:
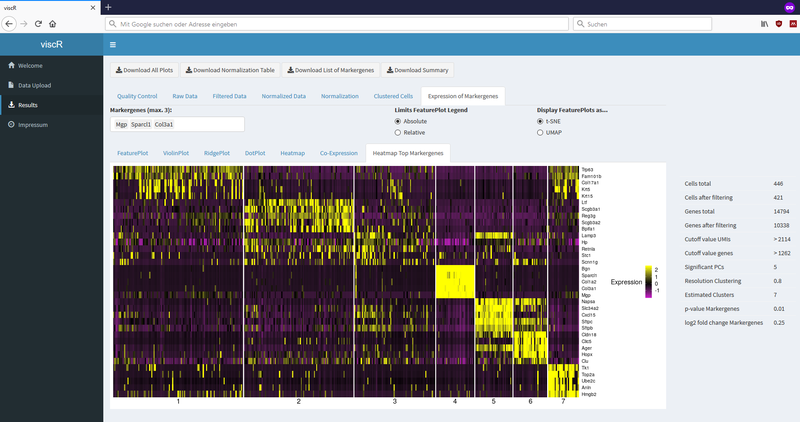
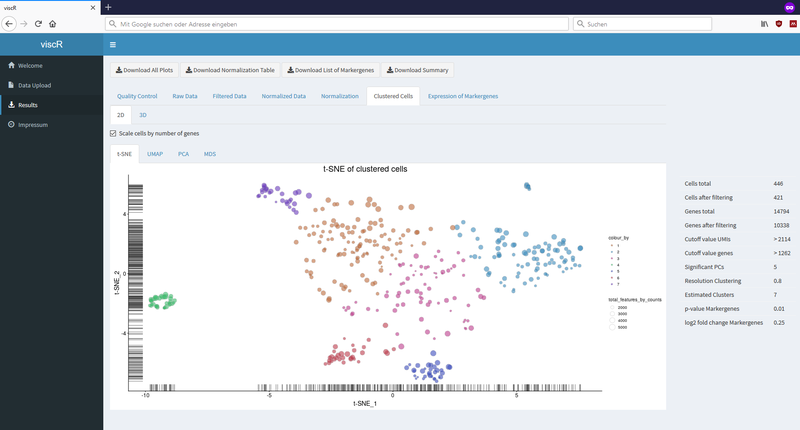
Exemplary screenshots: