Phylogenetic trees
As the main purpose of EDGAR is the estimation of orthologs among several genomes it is straightforward to infer phylogenetic knowledge from that orthology information.
Gontcharov et al. (2004) showed that the combination of several genes gives superior results when compared to phylogenetic trees derived from single genes like the popular 16s rRNA genes. In a phylogenetic analysis of 12 insect genomes Zdobnov and Bork (2007) recommend the use of all core genes of a set of genomes to maximize the sequence support for the phylogenetic tree. They propose a pipeline for the construction of reliable, core genome based phylogenetic trees which is used in EDGAR in a slightly adapted version.
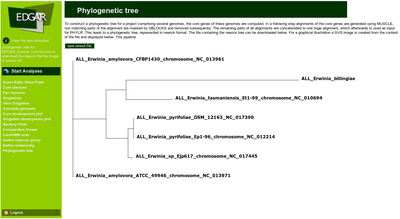
The pipeline calculates the core genome for a selection of genomes. Every set of orthologous genes found in all genomes is separately aligned using the multiple alignment tool MUSCLE (Edgar, 2004). The alignments are concatenated to one huge multiple alignment that can easily be hundreds of thousand of residues long. A distance matrix is calculated from this alignment and finally a phylogenetic tree is constructed based on this distance matrix using the Neighbor-Joining method. The two latter methods are used in the PHYLIP implementations by Felsenstein (1995). The Neighbor-Joining method was chosen as it is a heuristic approach with a very good computational efficiency, making it well suited for large datasets resulting from the core genome based tree construction.
