Software
The huge amounts of data acquired from PolyOmics technologies can only be handled with intensive bioinformatics support that has to provide an adequate data management, efficient data analysis algorithms, and user-friendly software applications. Over the last years, we developed a large scale software suite to substantially support researchers in the field of genome and metagenome, transcriptome, proteome, and metabolome data analysis. As all software applications follow a similar design pattern, experimental data as well as analysis results can be easily integrated to achieve higher-level evaluation and combined visualizations.
Forward-looking projects within our group focus on the development of bioinformatics analysis workflows for rapid and parallel data interpretation, which also includes hardware accelerated software implementations. Conveyor is our workflow-processing engine to rapidly deploy new bioinformatics data analysis pipelines utilizing either a compute cluster for distributed computing or multi-core servers for local execution. Using a workflow based approach, analyses can be designed using an intuitive graphical designer application. A number of ready-to-use processing steps for bioinformatics analyses already exist with a focus on sequence analysis and sequence annotation. Using the Conveyor2Go tool, an existing workflow can easily be converted into a standalone application.
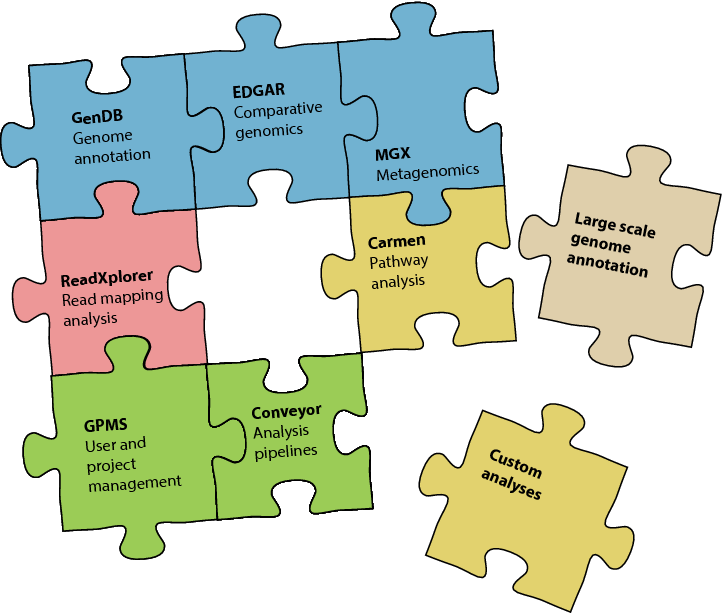
The whole collection of established software tools at hand provides the basis for efficient, parallel, and data driven processing that will be further improved and extended in the future to build a comprehensive bioinformatics technology platform for genome based systems biology.
