Project 1: Creating of a cell atlas of the liver fluke using single cell RNAseq
Classical transcriptome methods lack cellular resolution
RNA sequencing (RNAseq) can be used to measure transcription in all genes of an organism. Classical approaches use complex tissue samples composed of different cells for these methods. The disruption of the tissue creates a mixture and results therefore represent an average value of the transcription of a gene. Innovative methods such as "single-cell RNAseq" (sc-RNAseq) capture transcripts in single cells. The data set generated in this way impresses with higher resolution and allows more precise measurement of gene activity in individual cell types.
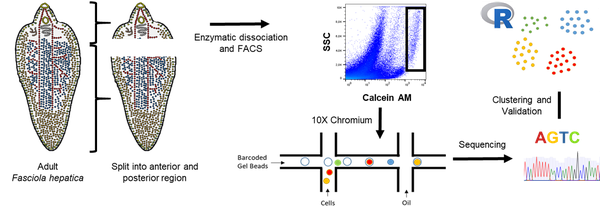
Creating a Cell Atlas for the Liver Fluke Fasciola hepatica
In this project, we are working on mapping cell types based on their transcription. This enables us to functionally analyze these cells and gain a deeper understanding of the biology of these parasites.
Besides its exciting applications in medical research, this innovative method provides a unique opportunity for studying new organisms. While classical model organisms such as mice, fruit flies, or the nematode Caenorhabditis elegans have been the focus of research for decades and data on various cell types are available, this isn't the case for many other organisms. High-throughput methods like sc-RNAseq offer the potential to acquire a vast amount of information.

Dissociated cells of the liver fluke A: Brightfield microscopy B: Fluorescence microscopy with live cell dye
Calcein
