AG Lochnit
copyright: private
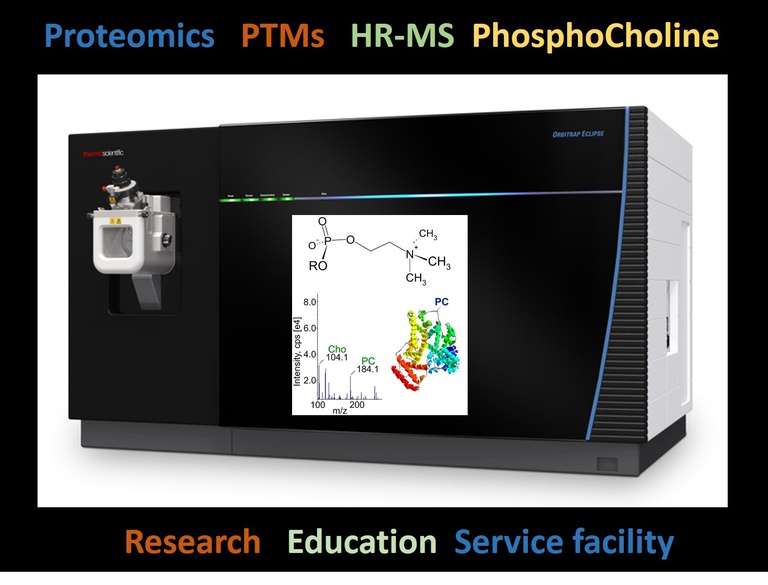
The Protein Analytics group has many years of experience in protein analytics. The methodological repertoire includes high-resolution electrospray mass spectrometry, 2D gel electrophoresis, densitometric analysis of proteomes and automated protein identification by MALDI-TOF mass spectrometry. In addition, the group has numerous other protein analytical methodologies at its disposal, such as N-terminal Edman sequencing and various liquid chromatographic separation methods.
The methods are generally available to interested scientific working groups within the framework of scientific cooperations. As a service facility, there are numerous collaborations with research groups from the Clinical Research Units, the Collaborative Research Centers and the Cluster of Excellence of the Department of Medicine at JLU Giessen as well as international graduate programs. In addition, there are numerous collaborations with working groups from other departments as well as universities on a national and international level.
In addition, Protein Analytics regularly offers workshops on methods of protein analytics for the training of young scientists.
Prof. Dr. Günter Lochnit is a member of the "International Giessen Graduate School for the Life Sciences (GGL)" and the HUPO initiative iMOP.
Research focus of the Protein Analytics group is method development in protein analytics with emphasis on automation and analysis of post-translational modifications. One focus is the analysis of post-translational modifications of pathogens, which enable the survival of the pathogen in the host. The model system is the free-living soil nematode Caenorhabditis elegans. For its investigation a C. elegans lab is connected to the protein analytics.
Other model systems can be found here.
A possible characterization of glycosphingolipids of nematodes we explain here.
RNA Interfernce (RNAi)
Immunomodulation: Phosphorylolcholine as immunomodulatory component
